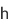Successful Construction of Artificial Cell Nuclei Inside of Fertilized Egg
A World First One Part of the Cell Nuclei Structure Formation Mechanism Elucidated
June 13, 2019
National Institute of Information and Communications Technology
Kindai University
Osaka University
1. Highlights of Study
- A world first, artificial creation of cell nuclei structures has been achieved by using microinjection of DNA-conjugated beads (with a diameter of about 3μm) into the cytoplasm of living fertilized mouse eggs
- The artificially created cell nuclei were confirmed as being the same as those created naturally by the fertilized eggs, with the formation of DNA nucleosomes with surrounding nuclear membranes
- The presence of DNA is shown to be the starting factor of the formation of cell nuclei inside of fertilized eggs, this knowledge is expected to contribute to the discovery of causes of infertility in the future
2. Content of the Research
3. About Publication
4. Background
5. Details
(1) Establishment of insertion method of DNA-conjugated beads to fertilized mouse eggs.
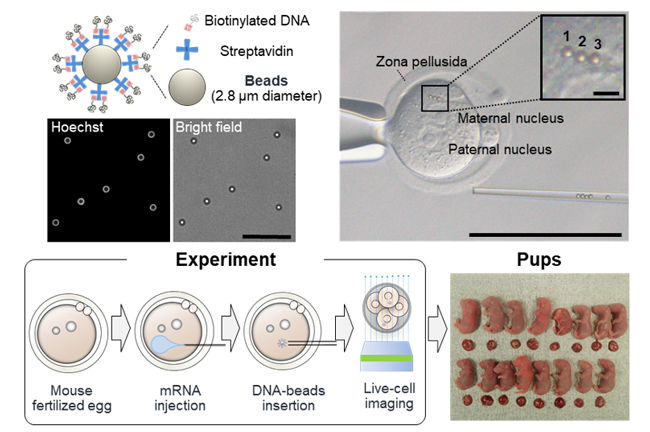
(2) Observation of histone accumulation and nucleosome structures surrounding the DNA-conjugated beads.
(3) Observation of the structure of the nuclear membrane and nuclear membrane pores surrounding the DNA-conjugated beads.
(4) Assessment of the transport ability of the artificial nuclei structures
6. Future Outlook
7. Glossary
Contact
Public Relations Office, General Affairs Department, Kindai University
Contact: Takahashi, Murao
Tel: +81-6-4307-3007 Fax: +81-6-6727-5288
E-mail:
















Press Office, Public Relations Department, National Institute of Information and Communications Technology
Contact: Hirota
Tel: +81-42-327-6923 Fax: +81-42-327-7587
E-mail:



















Graduate School of Frontier Biosciences, Osaka University
Prof. Yasushi Hiraoka
Tel: +81-6-6879-4620 Fax: +81-6-6879-4622
E-mail: